Longevity interventions are advancing through clinical trials, but predicting their long-term benefits remains a significant challenge. While many of these interventions show promise in preclinical studies and early-stage trials, their actual impact on health span over extended periods is difficult to determine due to the complexity of aging and the multifactorial nature of biological responses.
Epigenetic biomarkers, especially DNA methylation-based clocks, offer a promising solution by using DNA methylation patterns to track biological aging and assess intervention efficacy early. By assessing changes in epigenetic age in response to treatments, we can gain early indications of whether a given intervention is effectively slowing, reversing, or accelerating biological aging.
Epigenetics is tissue-specific. Acquiring large-scale skin epigenetic data is difficult due to the reliance on invasive procedures such as skin biopsies. Additionally, biopsy samples often contain a mixture of both the dermis and the epidermis. This variability introduces a significant challenge in epigenetic analysis, as differences in cell composition can lead to inconsistent and less reliable results. In contrast, the epidermis is a highly homogeneous tissue, composed of approximately 95% keratinocytes, making it an ideal source for consistent and reproducible epigenetic analysis.
At Mitra Bio, we have a non-invasive skin sampling method utilizing tape stripping to collect the top layer of the human epidermis. This approach allows us to obtain a highly homogeneous sample, consisting predominantly of keratinocytes, ensuring a more consistent and biologically relevant dataset for skin-specific epigenetic analysis.
We analyzed sequencing data obtained from tape-stripping using existing clocks and found that the error rates were high when applied to non-invasive skin samples. One notable example is the Horvath Skin and Blood clock which is a widely used epigenetic age estimator designed to predict chronological age based on DNA methylation patterns in both skin and blood tissues. When applied to blood samples, this clock has demonstrated a Mean Absolute Error (MAE) of approximately 2.5 years and R² 0.94, in skin samples, indicating a high level of accuracy in aligning predicted age with actual chronological age.
However, when applied to sequencing data from skin samples, the Horvath Skin and Blood Clock did not perform as well, still exhibited notable error rates, with a MAE of 8.95, and an R² value of 0.410. This indicates that existing epigenetic clocks may have limitations when applied to non-invasively collected skin samples, underscoring the need for skin-specific models optimized for tape-stripping data.
MitraClock
To address this limitation, we developed MitraClock, a skin-specific epigenetic clock trained on a large dataset of non-invasively collected skin samples from the face. Compared to existing clocks, MitraClock demonstrates superior accuracy in estimating epigenetic age in skin samples, achieving an MAE of 4.31 years and an R² value of 0.81—a significant improvement over the Horvath Skin and Blood Clock and other existing models. These results highlight the importance of using tissue-specific epigenetic clocks tailored for non-invasive sampling methods, offering a more precise and scalable approach for assessing biological skin age and tracking longevity interventions.
| Clock | Median Absolute Error (years) | Mean Absolute Error (years) | R² |
| MitraClock | 3.05 | 4.31 | 0.81 |
| Horvath Pan Tissue | 10.66 | 11.98 | 0.58 |
| Horvath Skin and Blood | 7.76 | 8.95 | 0.41 |
| Hannum | 10.43 | 11.99 | 0.08 |
| PhenoAge | 8.95 | 9.83 | 0.36 |
Table 1. Showing the error rate of MitraClock versus relevant other clocks when applied to sequencing data obtained using non-invasive skin sampling
MitraClock performance on photo-aged skin
To validate the clock read-out on producing biological aging, we used photoaging skin, a well-known factor for accelerated skin-aging. Using skin samples obtained non-invasively from the forehead (UV-exposed) and upper back (UV-protected), differential methylation analysis was performed.
Figure 1. Boxplot of the average beta value of global methylation profile of 75 participants samples from the forehead (UV-exposed) and upper back (UV-protected)
Biological Age Measurement
We used MitraClock to test the two sample types and observed that UV-protected samples were predicted to be 3.8 years younger than UV-exposed skin samples.
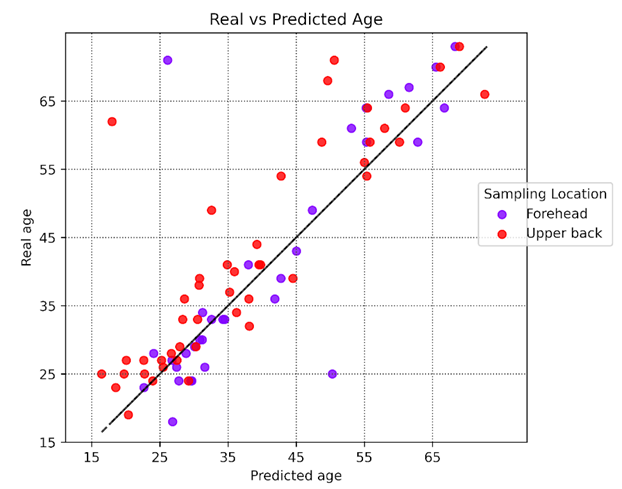
Figure 2. MitraClock representation of upper back and forehead skin samples for their real age (chronological age) and predicted age. The majority of upper back samples (UV-protected site) are younger than the forehead skin samples (UV-exposed sites)
Pace of Aging: UV-protected skin ages slower than UV-exposed skin on the face
In a matched cohort of forehead and upper back samples, MitraClock shows the pace of aging of upper back samples being 20% slower compared to the forehead samples.
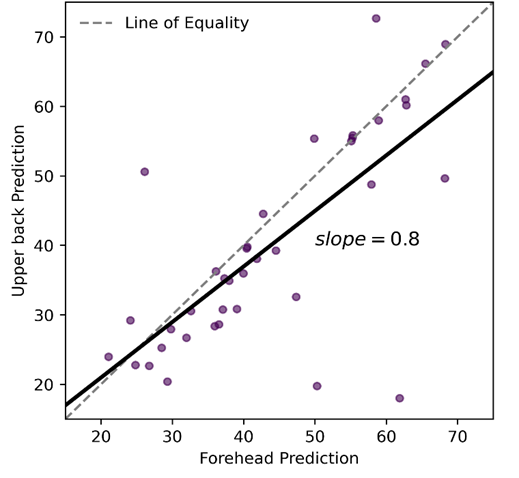
We ran differential methylation analysis to investigate the genes that are most impacted with photoaging.
- NRF2 – Activates antioxidant defense genes (e.g., SOD1, GCLC, HO-1) to mitigate UV-induced oxidative damage
- MMP3/MMP9 – Matrix metalloproteinases upregulated by UV, leading to collagen degradation and photoaging.
- DDB1/DDB2 – Critical for detecting UV-induced DNA damage as part of the nucleotide excision repair (NER) pathway.
In conclusion, MitraClock significantly outperforms established epigenetic clocks such as Horvath Pan Tissue, Horvath Skin and Blood, Hannum, and PhenoAge when applied to sequencing data derived from non-invasive skin samples. MitraClock achieved an R2 of 0.81, explaining 81% of the variance in biological age predictions, making it a highly reliable tool for skin aging studies and clinical applications.
References
Horvath, Steve et al. “Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies.” Aging vol. 10,7 (2018): 1758-1775. doi:10.18632/aging.101508
Banila, Cristiana et al. “A noninvasive method for whole-genome skin methylome profiling”, British Journal of Dermatology, Volume 189, Issue 6, December 2023, Pages 750–759,
Mitra Bio has developed a non-invasive skin diagnostics platform that measures biological skin aging. Since skin aging is a highly visible biomarker, tackling it could significantly advance the field of longevity.
If you’re developing longevity ingredients and want to track efficacy using DNA methylation age clocks, let’s connect.


